In silico antibody discovery and optimization: How computational tools are reshaping the workflow
AI-enabled and ML-powered methods are enabling faster identification and engineering of high-quality therapeutic antibody leads
The field of antibody discovery is experiencing a transformative shift through the integration of cutting-edge artificial intelligence (AI) and machine learning (ML) technologies. Traditional methods of antibody discovery are often costly, time-consuming, and labor intensive. Typically, they involve multiple rounds of optimization and are inherently limited in their ability to fully explore antibody sequence diversity. Now, powerful AI and ML computational techniques are dramatically accelerating and refining antibody discovery and development workflows, helping to increase the speed and probability of identifying high-quality therapeutic antibody candidates.1
By combining advanced AI and ML computational approaches with wet-lab validation, high-quality antibody leads can be identified and optimized faster. These techniques are synergistic – computational methods inform and guide experimental work while experimental data confirms in silico predictions and enables further training of models.1,2 AI and ML methods are being used to rapidly humanize leads, support discovery through precise variant design, and develop focused screening libraries, reducing timelines, costs, and risks for downstream developability and manufacturing challenges. This combined approach ensures that the best antibody candidates are identified quickly and with greater confidence.
Improving efficiency of traditional discovery workflows through in silico clone selection, developability prediction, and optimization
Generative AI approaches, particularly protein language models3 and diffusion models4, are now enabling researchers to create focused libraries enriched with candidates predicted to bind specific epitopes or exhibit desired biophysical traits, moving beyond traditional random screening. Two primary generative approaches have emerged: one focused on generating three-dimensional antibody structures, and the other on generating amino acid sequences (with hybrids or combinations of both approaches actively investigated). These models can be conditioned on specific epitopes or antigens, thereby generating novel proteins and antibodies in a context-dependent manner. While these AI-driven methods require substantial data and computational resources for training, the created models also have the potential to be reused and fine-tuned for related tasks.
Despite their promise, current generative AI methods for de novo antibody design face limitations, including low hit rates (<1%) and suboptimal affinities, necessitating large-scale library screens to identify viable candidates. While advancements in immune receptor data and computational techniques will improve these outcomes over time, AI and ML’s greatest near-term impact likely lies in augmenting—not replacing—traditional workflows.
Most approved antibodies still originate from in vivo platforms primarily through immunization of wild-type or humanized mice (such as ATX-Gx™) , where natural immune selection ensures high affinity and developability. An alternative workflow leverages synthetic human antibody libraries displayed on the surface of yeast, mammalian cells, or phage for selection. While effective, these workflows require additional resources to identify and characterize binders with desired properties, typically screening large numbers of clones. In silico tools, including AI and ML, have the potential to significantly streamline key steps in these processes, such as clone selection, developability screening, and affinity maturation by prioritizing candidates predicted to have optimal properties, reducing reliance on labor-intensive experimental iterations.
During the discovery phase, in silico tools including AI and ML can significantly improve the efficiency of clone selection and prioritization. Traditional in vivo discovery workflows like hybridoma or single B cell sorting require expression and screening of large numbers of putative binders, which are often dominated by a limited number of expanded clonotypes, reducing the overall diversity. Instead, deep sequencing of the immune repertoire after immunization and enrichment, followed by in silico analysis to identify antigen-experienced clones can be used to predict a set of likely binders designed to maximize sequence and epitopic diversity (see Figure 1).5
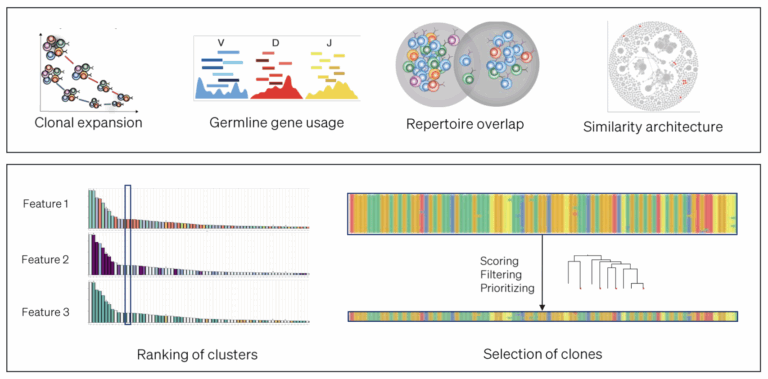
In silico tools can also be applied during discovery to eliminate clones predicted to have development liabilities such as instability, aggregation propensity, and polyreactivity. Analysis of antibody drug-like properties is currently done empirically, often late in the characterization funnel. ML-trained models based on experimentally determined developability data can be used to predict protein liabilities based on sequence or predicted structure, enabling efficient prioritization for drug-like properties earlier in the discovery cascade. These predictive models enable testing of a smaller number of potential clones with a higher hit rate of binders that match the target product profile.6,7
AI-enabled and ML-powered antibody discovery
Download our poster to discover how AI and ML technologies are applied to accelerate antibody discovery and are able to generate high quality functional leads
Current AI and ML in silico approaches are potentially most impactful for antibody optimization, where they are utilized to fine-tune properties like affinity, specificity, and developability. Instead of engineering optimized antibody candidates using complex yeast and phage display mutagenesis libraries, AI and ML methods focus on targeted expression and characterization of individual variants through a controlled “design, build, test” feedback loop, often also called ‘lab-in-the-loop’. 8,9, 10 These advanced computational techniques enable more precise and efficient variant selection, allowing researchers to identify high-quality therapeutic candidates with fewer experimental iterations, bridging the gap between computational innovation and practical application in antibody discovery.
Proprietary AI/ML workflows in action at Alloy Therapeutics
Here at Alloy, we take a precision-driven approach to generating, retrieving, and evaluating antibodies with optimal therapeutic properties. Leveraging our ATX-Gx™ humanized mice platform, mAbForge (rapid express-test workflow), advanced in vitro functional testing, and a proprietary suite of AI and ML tools, we deliver data-informed, efficient, and accelerated antibody discovery and development services. Our AI and ML capabilities include advanced antibody data visualization, computationally optimized antibody selection from in vivo repertoires, digital mAb clonotype expansion, AI-enabled variant design, in silico library development, and customizable machine learning and deep-learning models to predict antibody liabilities — all designed to streamline and enhance therapeutic antibody development. These capabilities fuel our ability to generate high-quality, diverse, and developable antibodies with desired binding affinities, improved stability, enhanced specificity, species cross-reactivity, humanized sequences, and pH-dependent binding.
We are committed to using AI and ML to drive innovation in antibody discovery, developing and applying cutting-edge computational tools to address unmet needs. By integrating AI and ML with mAbForge, our rapid-test proprietary workflow, we are well-positioned to discover and develop high-quality antibody therapeutics with improved efficiency and success rates.
Are you ready to explore how Alloy Therapeutics’ Antibody Discovery Services can help you rapidly identify and optimize your next therapeutic antibody candidate? Contact us to discuss your project.